News
Exploring The Microbiomes Of Fermented Foods
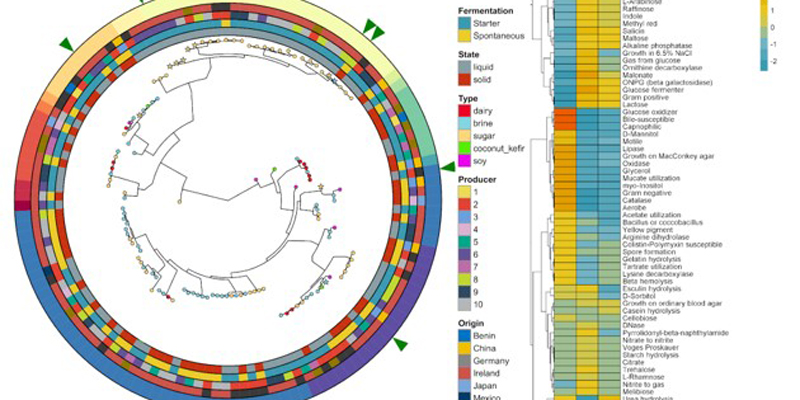
Exploring The Microbiomes Of Fermented Foods
Contributed by John Leech
Our new study investigated 58 fermented foods from 8 countries and identified factors driving the microbial composition of these foods and the potential functional benefits associated with these populations.
Using the latest in DNA sequencing technology, we uncovered the taxonomic and functional profile of these 58 foods. The main conclusion of the study was that food substrate is the primary factor that drives microbial diversity within the foods. Country of origin, specific producer of the food and the use of starter cultures versus spontaneous fermentation had a smaller impact. We examined three major substrates; brine, dairy and sugar foods. Dairy foods contained the lowest diversity while brine and sugar foods had similar diversity profiles to each other.
The taxonomic profile of the 58 foods revealed many familiar fermented food species. However, many novel taxa (not previously discovered in these foods) were also uncovered, including 10 species that are new to science. Several fermented foods were analysed with DNA sequencing techniques for the first time, including boza, lacto-fermented cucumber and beet kvass.
Functional profiling of microbiomes allows the examination of other qualities aside from taxonomy. In this study, the general functional profile, bacteriocin profile, carbohydrate utilisation profile and anti-microbial resistance profile were characterised. The general functional profile of these foods provides information on what specific functions are contained within a microbial profile. The bacteriocin profile sheds light on some of the selective pressures that exist within a specific food microbiome and will also help with future studies designed to uncover bacteriocin producers from fermented foods. Many of the foods contained the functional potential to produce bacteriocins that are affective against food-borne pathogens such as Listeria.
We also explored he anti-microbial profile of fermented foods for the first time. Most of the foods had less anti-microbial resistance genes than human gut microbiome samples, but a small number of foods did contain more of these genes. This study has established a baseline of anti-microbial resistance in fermented foods.
Fermented foods are re-emerging as popular food choices in some cultures, driven by the purported health benefits of consuming live fermented foods. The 58 foods in this study were screened for genes that have been shown to be vital for health benefits in previously known probiotic taxa. Non-fermented foods were also examined for comparative purposes. Overall, fermented foods contained many more potentially health promoting genes than non-fermented foods. Foods containing sugar, such as kombucha and water kefir, generally had the highest health promoting potential. This is the first time this type of analyses has been deployed.
The results of this study open up many more questions for fermented food research, including the need to examine the health promoting potential of several candidate foods that exhibited strong results from the genetic health-promoting screening.
Original article:
John Leech et al (2020) mSystems DOI: 10.1128/mSystems.00522-20
.png)





